In [1]:
%matplotlib inline
%load_ext autoreload
%autoreload 2
In [2]:
import scanpy as sc
import random
from unicoord import scu
from unicoord.visualization import draw_loss_curves
import torch
from line_profiler import LineProfiler
In [3]:
sc.settings.verbosity = 3 # verbosity: errors (0), warnings (1), info (2), hints (3)
sc.logging.print_header()
# sc.settings.set_figure_params(dpi=80, facecolor='white')
sc.settings.set_figure_params(vector_friendly=False)
load hECA data¶
In [ ]:
adata = sc.read_h5ad(r"F:\h5ad\hECA_eachCT2000_pcGenes.h5ad")
sc.pp.normalize_total(adata, target_sum=1e4 ,exclude_highly_expressed= True)
sc.pp.log1p(adata)
adata
model and training¶
In [ ]:
scu.model_unicoord_in_adata(adata, n_diff=0, n_clus=[], n_cont=20,
obs_fitting = ['seq_tech', 'organ','cell_type'])
In [30]:
scu.train_unicoord_in_adata(adata, epochs=2, slot = 'cur', chunk_size=20000)
In [31]:
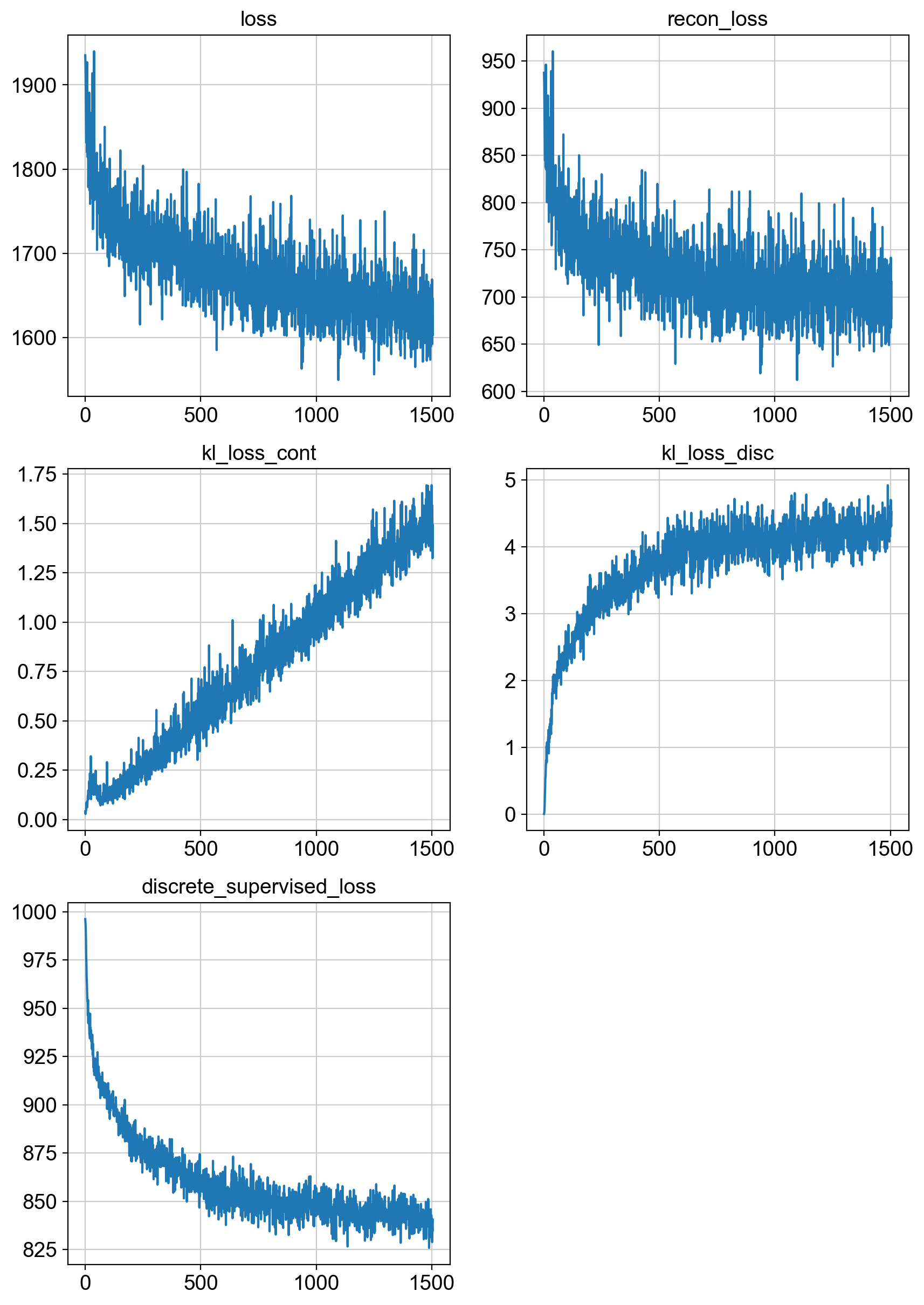
fig = draw_loss_curves(adata.uns['unc_stuffs']['trainer'].losses)
# if save_figs:
# fig.savefig(os.path.join(savePath, 'img', 'fig1_lossCurves.png'))
fig.show()
In [32]:
scu.embed_unicoord_in_adata(adata, only_sup=False)
In [33]:
adata.obsm['unicoord'].shape
Out[33]:
In [34]:
sc.pp.neighbors(adata, use_rep='unicoord')
In [35]:
sc.tl.leiden(adata, resolution=0.5)
In [36]:
sc.tl.umap(adata)
In [38]:
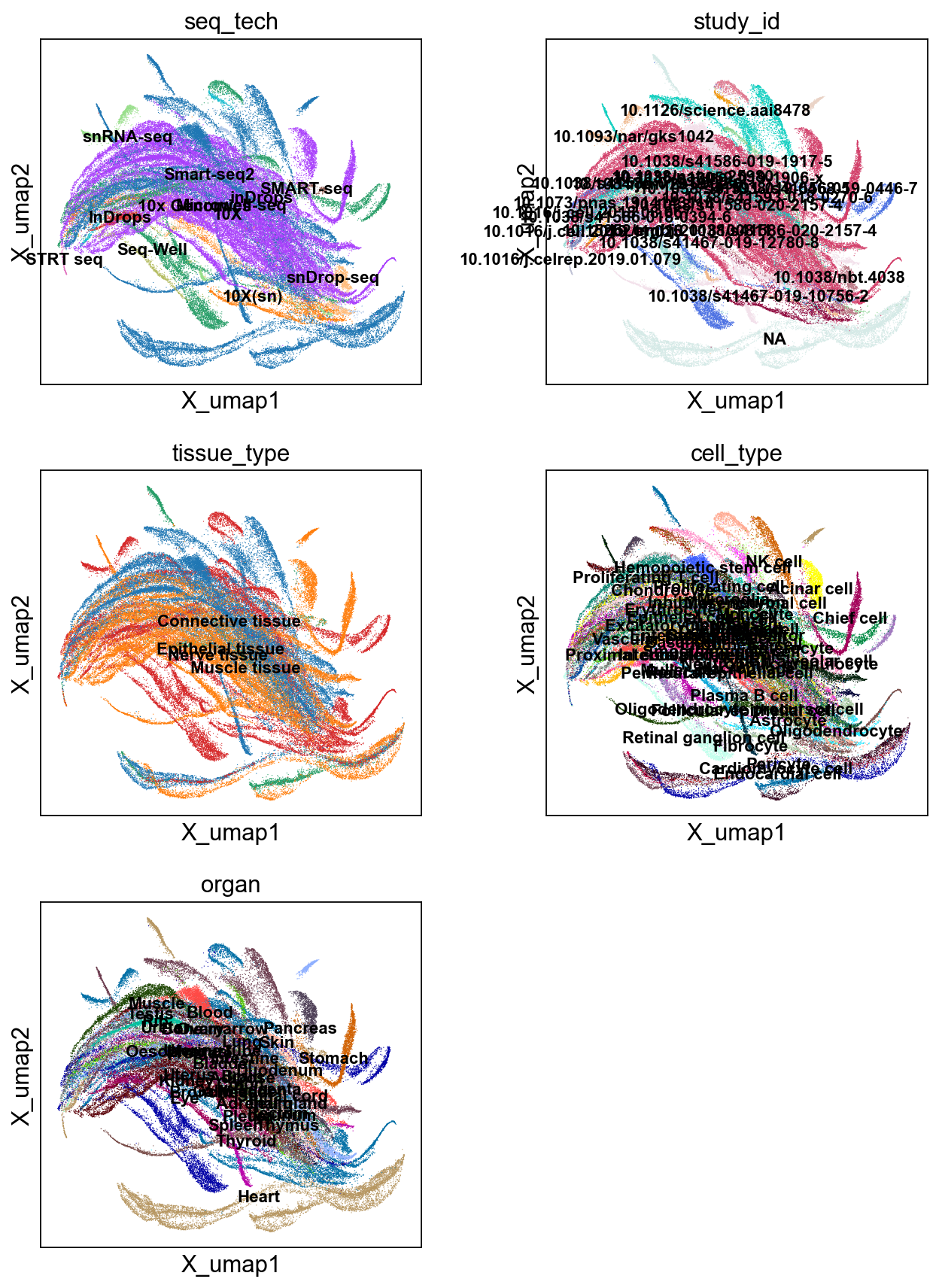
sc.pl.embedding(adata, 'X_umap',legend_loc='on data', legend_fontsize=10,
color=['seq_tech', 'study_id', 'tissue_type','cell_type','organ'], ncols=2)
predict test set¶
In [42]:
bdata = adata[~adata.obs.unc_training,:].copy()
bdata
Out[42]:
In [43]:
scu.predcit_unicoord_in_adata(bdata, adata)
In [44]:
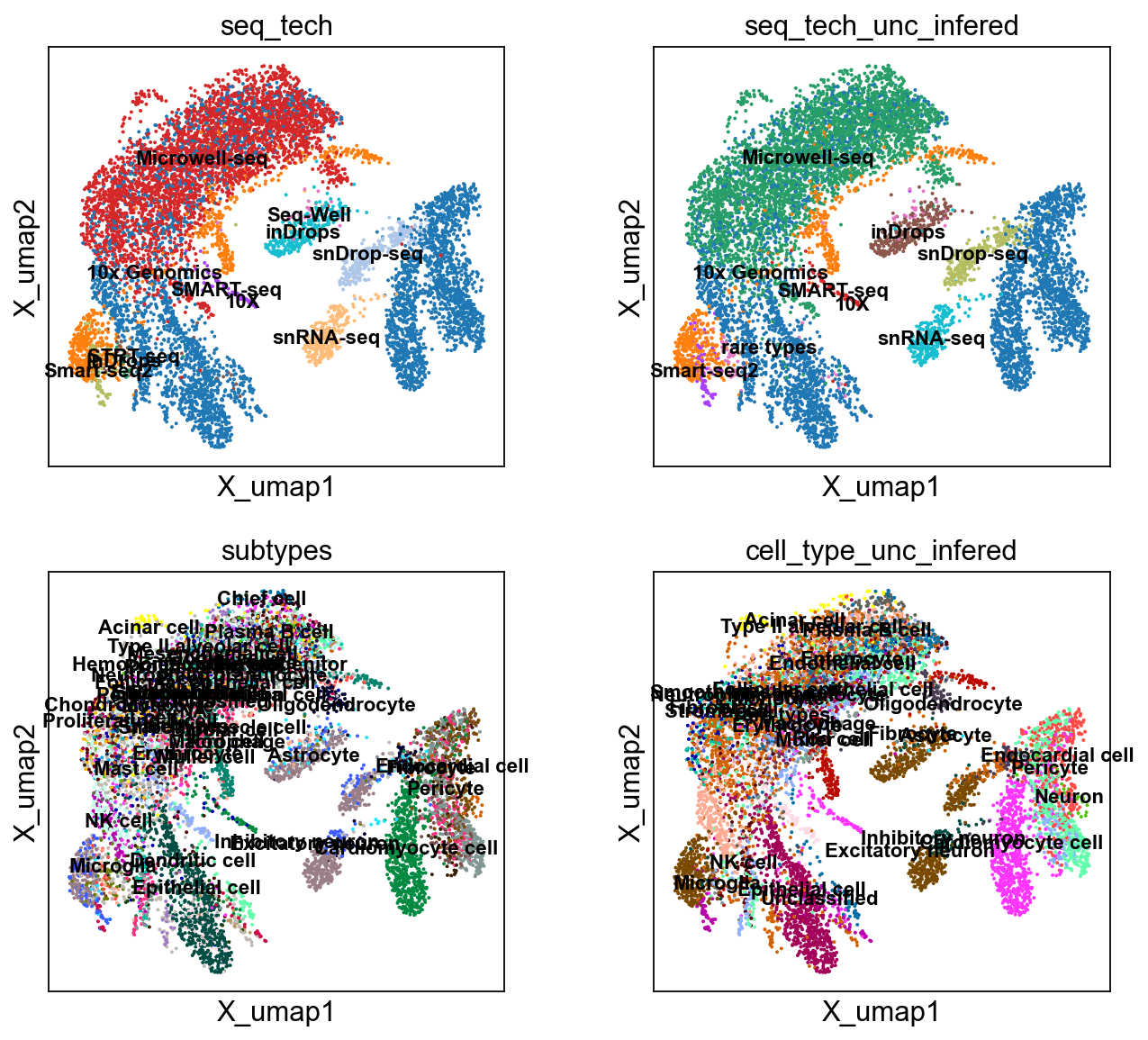
sc.pl.embedding(bdata, 'X_umap',legend_loc='on data', +
color= ['seq_tech', 'seq_tech_unc_infered',
'cell_type', 'cell_type_unc_infered',
'organ','organ_infered'], ncols=2)
predict LUAD data¶
In [52]:
cdata = sc.read_h5ad(r'D:\hECA\Lung_cancer.pp.h5ad')
In [53]:
cdata
Out[53]:
In [54]:
sc.pl.embedding(cdata, 'X_umap',legend_loc='on data', legend_fontsize=10,
color=['leiden','Sample','Cell_type.refined','Cell_subtype'], ncols=2)
In [55]:
cdata = cdata.raw.to_adata()
sc.pp.normalize_total(cdata, target_sum=1e4 ,exclude_highly_expressed= True)
sc.pp.log1p(cdata)
cdata
Out[55]:
In [56]:
scu.predcit_unicoord_in_adata(cdata, adata)
In [64]:
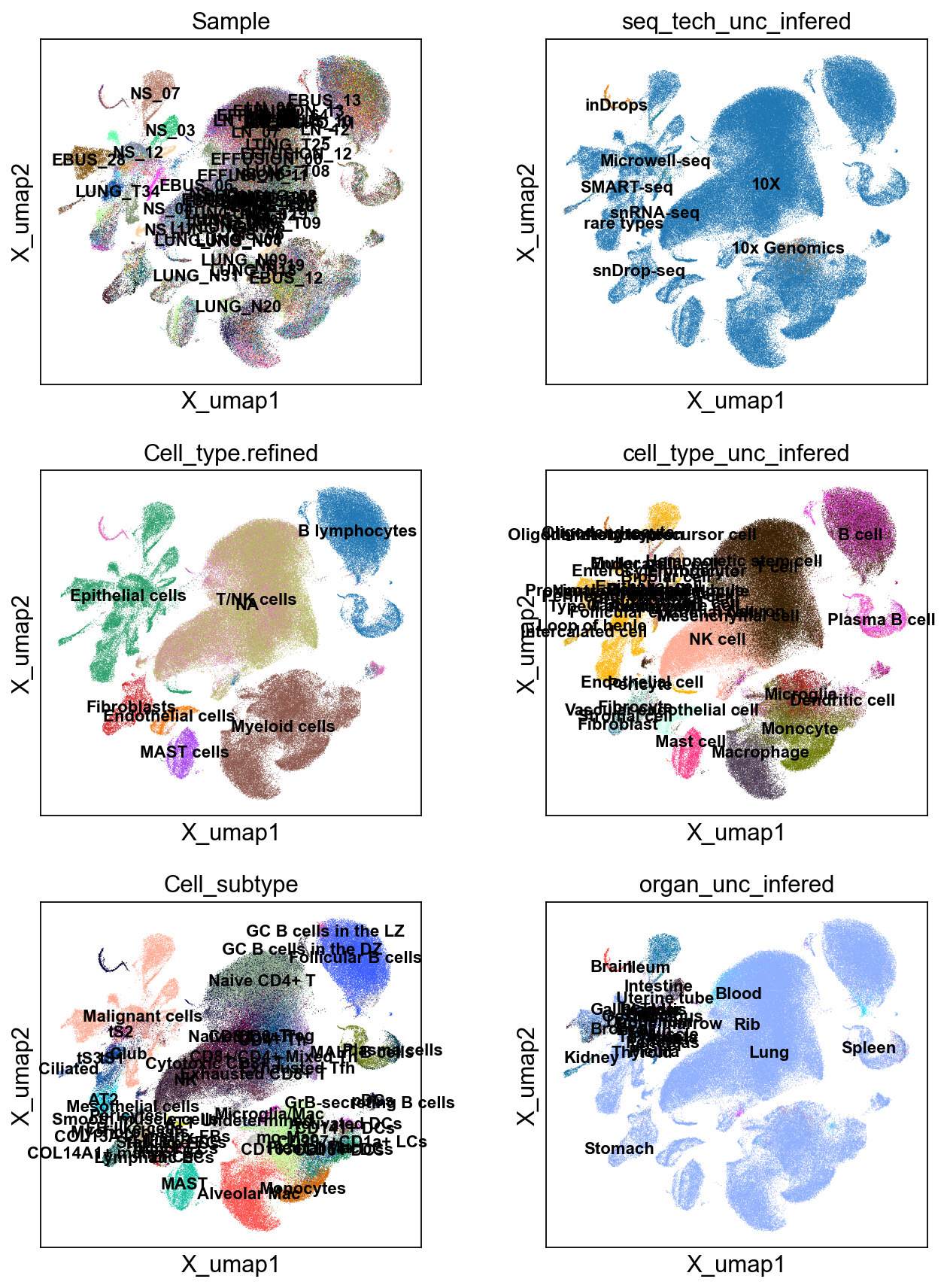
sc.pl.embedding(cdata, 'X_umap',legend_loc='on data', legend_fontsize=10,
color= ['Sample', 'seq_tech_unc_infered',
'Cell_type.refined', 'cell_type_unc_infered',
'Cell_subtype', 'organ_unc_infered'], ncols=2)
In [46]:
confusion_mtx = pd.read_csv('./table.csv', index_col=0)
In [59]:
confusion_mtx.columns
Out[59]:
In [63]:
cdata.obs.groupby(['cell_type_unc_infered','Cell_type.refined']).size().unstack()
Out[63]:
In [61]:
confusion_matrix = cdata.obs.groupby(['cell_type_unc_infered','Cell_subtype']).size().unstack().loc[:,list(confusion_mtx.columns)[:-1]]
confusion_matrix.to_csv('./hECA_pred_LUAD.csv')
In [102]:
ct_mapping = {"Epithelial cell" : "Epithelial cells",
"Type II alveolar cell" : "Epithelial cells",
"Goblet cell" : "Epithelial cells",
"Basal cell" : "Epithelial cells",
"Enterocyte progenitor" : "Epithelial cells",
"Vascular endothelial cell" : "Endothelial cells",
"Stromal cell" :"Fibroblasts",
"Fibroblast" : "Fibroblasts",
"Fibrocyte" : "Fibroblasts",
"Smooth muscle cell" : "Fibroblasts",
"Dendritic cell" : "Myeloid cells",
"Macrophage" : "Myeloid cells",
"Monocyte" : "Myeloid cells",
"Microglia" : "Myeloid cells",
"Mast cell" : "MAST cells",
"NK cell" : "T/NK cells",
"T cell" : "T/NK cells",
"B cell" : "B lymphocytes",
"Plasma B cell" : "B lymphocytes",
"Acinar cell" : 'other',
"Astrocyte" : 'other',
"Bipolar cell" : 'other',
"Cardiomyocyte cell" : 'other',
"Chief cell" : 'other',
"Endocardial cell" : 'other',
"Endothelial cell" : 'other',
"Erythrocyte" : 'other',
"Follicular epithelial cell" : 'other',
"Hemopoietic stem cell" : 'other',
"Inhibitory neuron" : 'other',
"Intercalated cell" : 'other',
"Loop of henle" : 'other',
"Mesenchymal cell" : 'other',
"Muller cell" : 'other',
"Neutrophilic granulocyte" : 'other',
"Neuron" : 'other',
"Oligodendrocyte" : 'other',
"Oligodendrocyte precursor cell" : 'other',
"Proliferating T cell" : 'other',
"Proximal convoluted tubule" : 'other',
"Pericyte" : 'other',
"Perineural epithelial cell" : 'other'}
In [114]:
ct1 = cdata.obs['Cell_type.refined']
ct2 = pd.Series([ct_mapping[c] if c in ct_mapping else "rare types" \
for c in cdata.obs['cell_type_unc_infered']])
ct1 = ct1[list(~cdata.obs['Cell_type.refined'].isin(['Undetermined','NA']))]
ct2 = ct2[list(~cdata.obs['Cell_type.refined'].isin(['Undetermined','NA']))]
In [115]:
from sklearn.metrics import accuracy_score
In [116]:
accuracy_score(ct1, ct2)
Out[116]:
In [117]:
ct2.index = ct1.index
a = pd.concat([ct1,ct2], axis=1)
a.columns = ['LUAD_label', 'pred_label']
In [118]:
a.groupby(['LUAD_label', 'pred_label']).size().unstack()
Out[118]:
In [119]:
confusion_mtx = pd.read_csv('./hECA_pred_LUAD.csv', index_col=0)
In [121]:
sns.set(rc = {'figure.figsize':(15,10)})
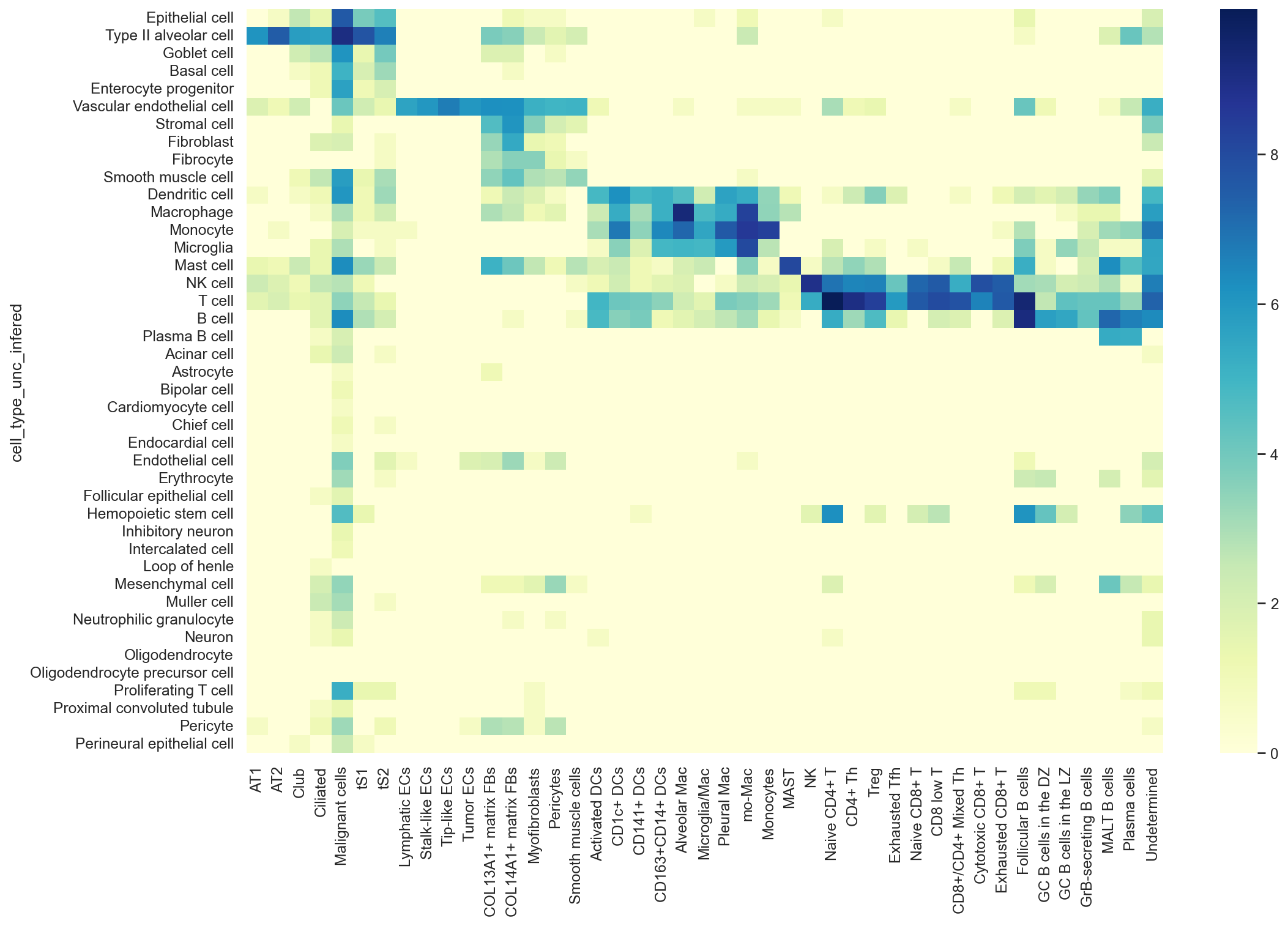
hcl = sns.heatmap(np.log(np.add(confusion_mtx, 1)),
cmap="YlGnBu",
xticklabels = True).figure.savefig('../UniCoord/hECA_pred_LUAD.pdf', bbox_inches="tight", dpi = 200)
In [122]:
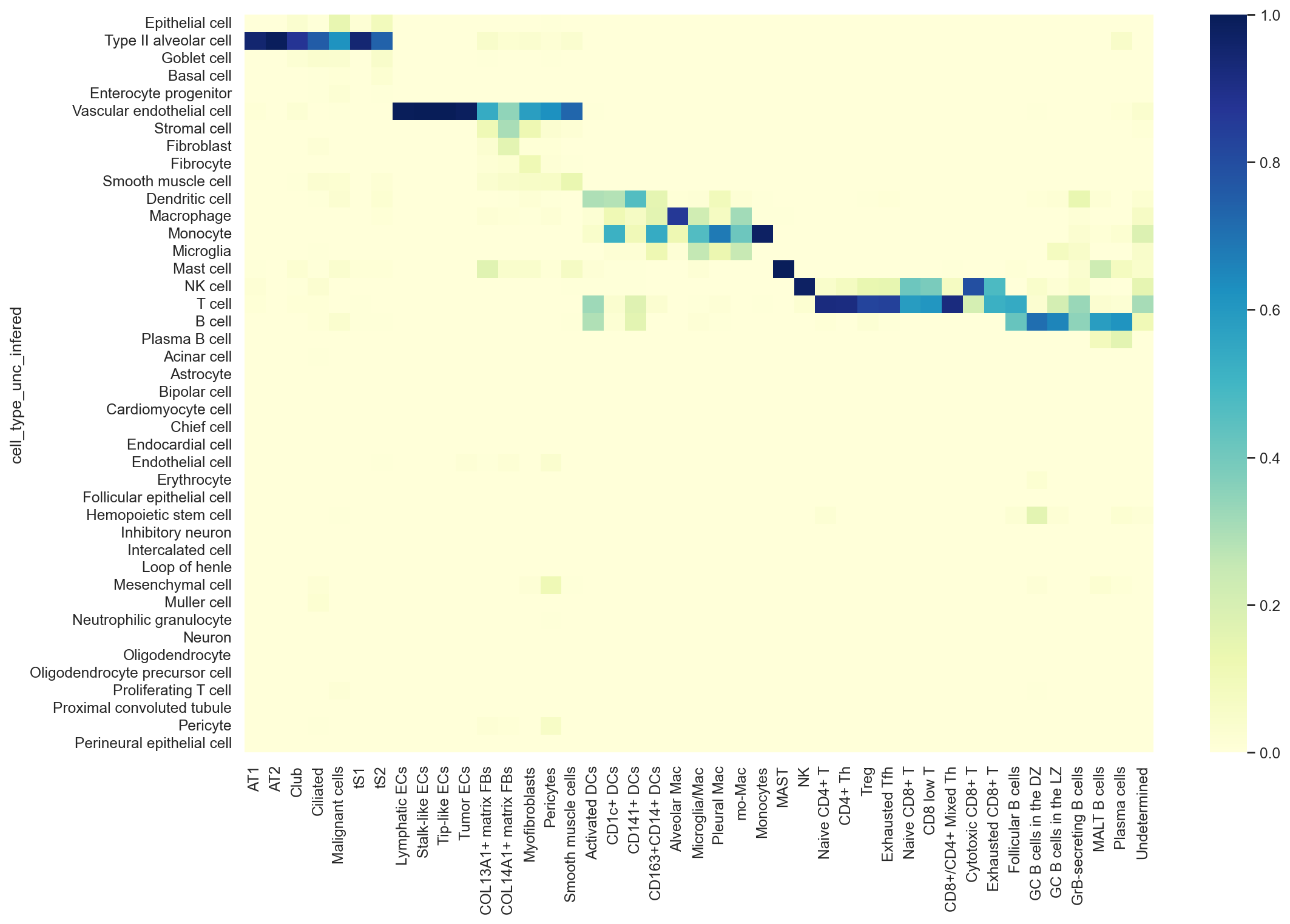
hcl = sns.heatmap(confusion_mtx/np.sum(confusion_mtx, axis=0),
cmap="YlGnBu",
xticklabels = True).figure.savefig('../UniCoord/hECA_pred_LUAD_ratio.pdf', bbox_inches="tight", dpi = 200)
In [123]:
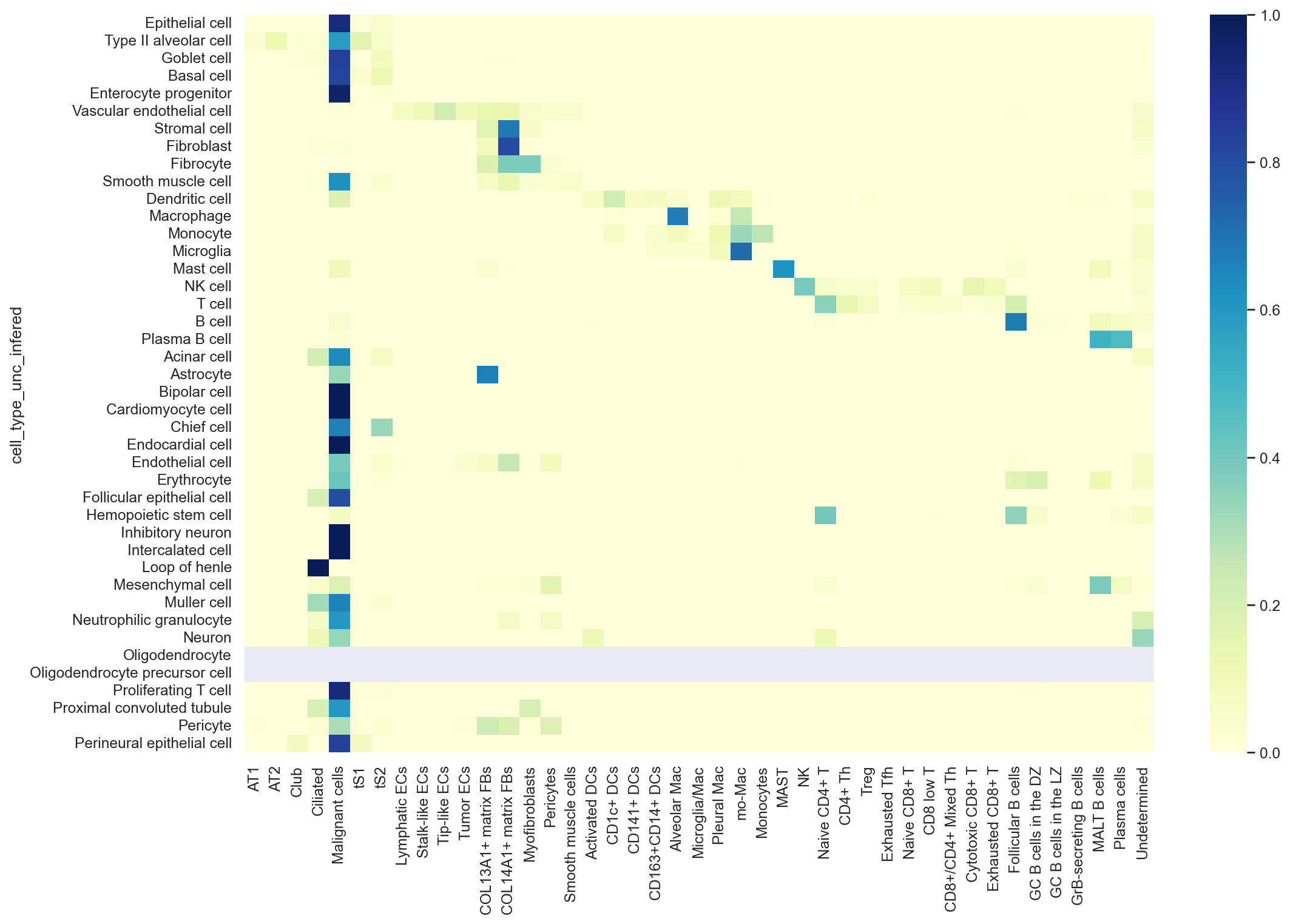
hcl = sns.heatmap((confusion_mtx.T/np.sum(confusion_mtx, axis=1)).T,
cmap="YlGnBu",
xticklabels = True)
predict lung ECA data¶
In [124]:
cdata = sc.read_h5ad(r'D:\hECA\Lung.Adult.pp.h5ad')
In [129]:
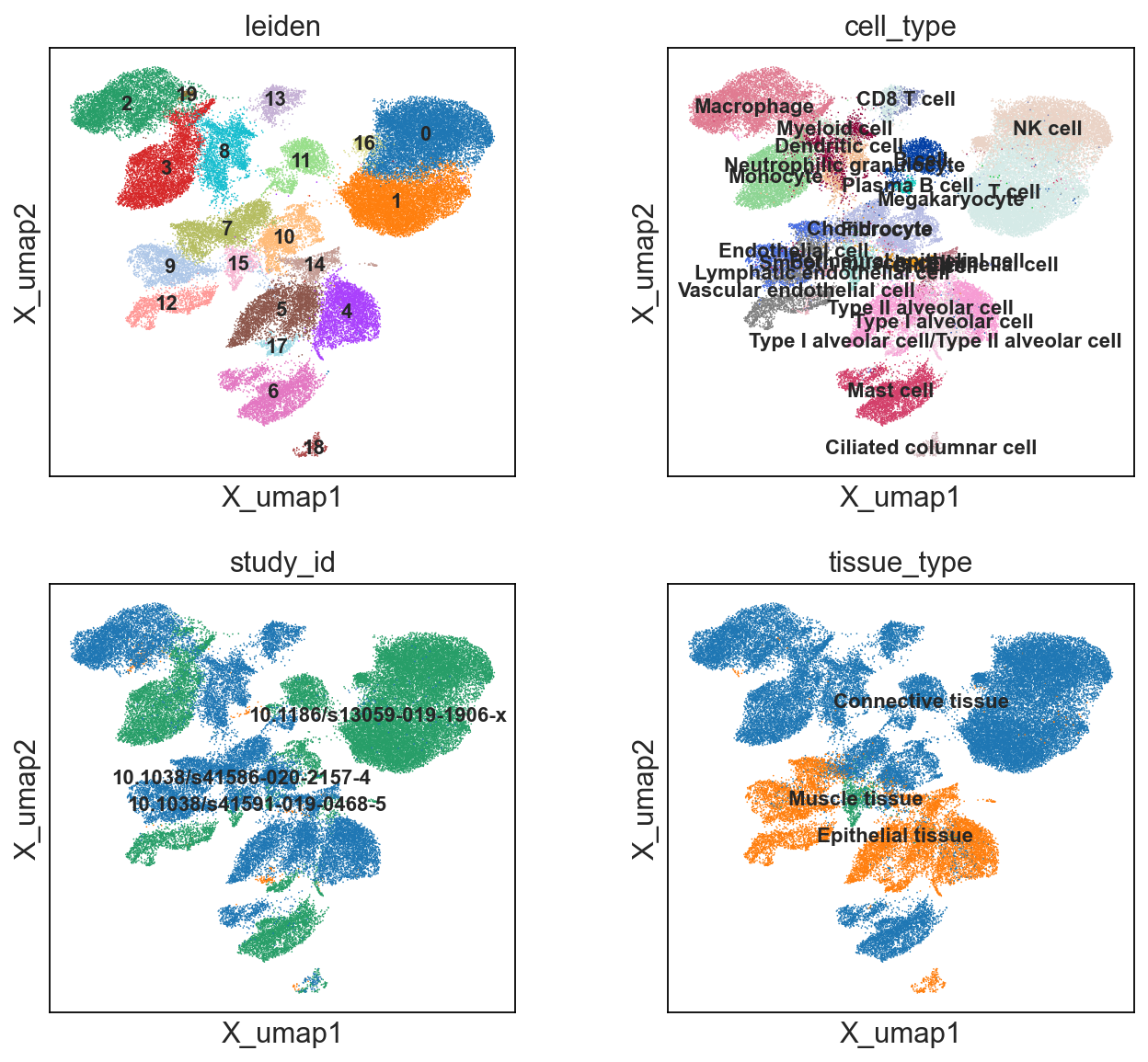
sc.pl.embedding(cdata, 'X_umap',legend_loc='on data', legend_fontsize=10,
color=['leiden','cell_type','study_id','tissue_type'], ncols=2)
In [126]:
cdata = cdata.raw.to_adata()
sc.pp.normalize_total(cdata, target_sum=1e4 ,exclude_highly_expressed= True)
sc.pp.log1p(cdata)
cdata
Out[126]:
In [127]:
scu.predcit_unicoord_in_adata(cdata, adata)
In [131]:
sc.pl.embedding(cdata, 'X_umap',legend_loc='on data', legend_fontsize=10,
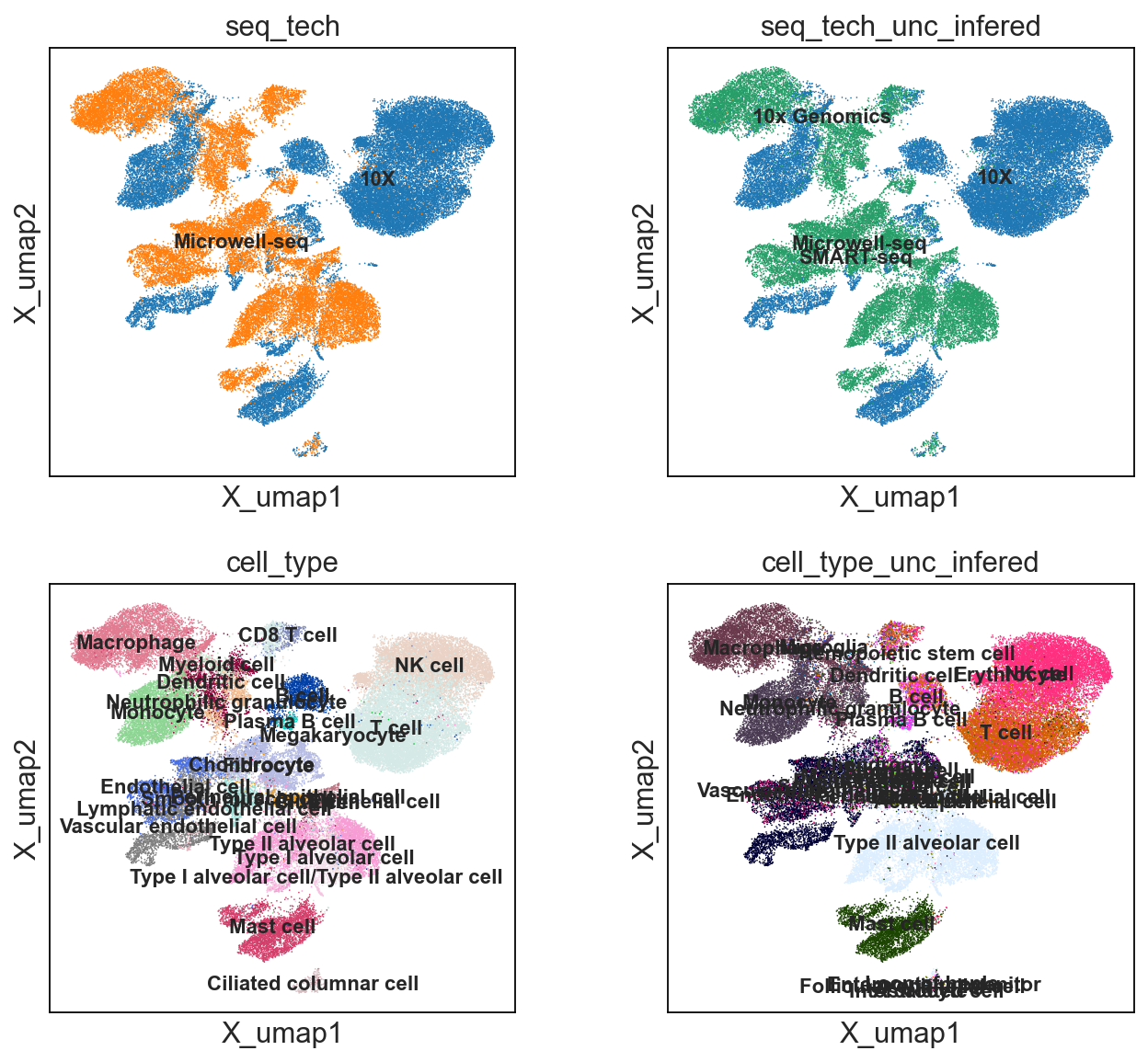
color= ['seq_tech', 'seq_tech_unc_infered', 'cell_type', 'cell_type_unc_infered'], ncols=2)
In [135]:
ct1_mapping = {"Epithelial cell" : "Epithelial cells",
"Type II alveolar cell" : "Epithelial cells",
"Goblet cell" : "Epithelial cells",
"Basal cell" : "Epithelial cells",
"Enterocyte progenitor" : "Epithelial cells",
"Vascular endothelial cell" : "Endothelial cells",
"Stromal cell" :"Fibroblasts",
"Fibroblast" : "Fibroblasts",
"Fibrocyte" : "Fibroblasts",
"Smooth muscle cell" : "Fibroblasts",
"Dendritic cell" : "Myeloid cells",
"Macrophage" : "Myeloid cells",
"Monocyte" : "Myeloid cells",
"Microglia" : "Myeloid cells",
"Mast cell" : "MAST cells",
"NK cell" : "T/NK cells",
"T cell" : "T/NK cells",
"B cell" : "B lymphocytes",
"Plasma B cell" : "B lymphocytes",
"Acinar cell" : 'other',
"Astrocyte" : 'other',
"Bipolar cell" : 'other',
"Cardiomyocyte cell" : 'other',
"Chief cell" : 'other',
"Endocardial cell" : 'other',
"Endothelial cell" : 'other',
"Erythrocyte" : 'other',
"Follicular epithelial cell" : 'other',
"Hemopoietic stem cell" : 'other',
"Inhibitory neuron" : 'other',
"Intercalated cell" : 'other',
"Loop of henle" : 'other',
"Mesenchymal cell" : 'other',
"Muller cell" : 'other',
"Neutrophilic granulocyte" : 'other',
"Neuron" : 'other',
"Oligodendrocyte" : 'other',
"Oligodendrocyte precursor cell" : 'other',
"Proliferating T cell" : 'other',
"Proximal convoluted tubule" : 'other',
"Pericyte" : 'other',
"Perineural epithelial cell" : 'other'}
In [136]:
ct2_mapping = {"Type I alveolar cell" : "Epithelial cells",
"Type I alveolar cell/Type II alveolar cell" : "Epithelial cells",
"Type II alveolar cell" : "Epithelial cells",
"Club cell" : "Epithelial cells",
"Ciliated columnar cell" : "Epithelial cells",
"Perineural epithelial cell" : "Epithelial cells",
"Epithelial cell" : "Epithelial cells",
"Lymphatic endothelial cell" : "Endothelial cells",
"Vascular endothelial cell" : "Endothelial cells",
"Endothelial cell" : "Endothelial cells",
"Fibrocyte" : "Fibroblasts",
"Smooth muscle cell" : "Fibroblasts",
"Dendritic cell" : "Myeloid cells",
"Macrophage" : "Myeloid cells",
"Monocyte" : "Myeloid cells",
"Neutrophilic granulocyte" : "Myeloid cells",
"Myeloid cell" : "Myeloid cells",
"Mast cell" : "MAST cells",
"NK cell" : "T/NK cells",
"T cell" : "T/NK cells",
"CD8 T cell" : "T/NK cells",
"B cell" : "B lymphocytes",
"Plasma B cell" : "B lymphocytes",
"Chondrocyte" : "rare types",
"Megakaryocyte" : "rare types"}
In [137]:
ct1 = [ct1_mapping[c] if c in ct1_mapping else "rare types"
for c in cdata.obs['cell_type_unc_infered']]
ct2 = [ct2_mapping[c] if c in ct2_mapping else "rare types"
for c in cdata.obs['cell_type']]
In [143]:
from sklearn.metrics import f1_score
In [145]:
f1_score(ct1, ct2, average='micro')
Out[145]:
In [147]:
f1_score(ct1, ct2, average='macro')
Out[147]:
In [146]:
accuracy_score(ct1, ct2)
Out[146]:
In [140]:
a = pd.DataFrame([ct1,ct2]).T
a.columns = ['pred','true']
In [141]:
a.groupby(['pred','true']).size().unstack()
Out[141]: