In [1]:
%matplotlib inline
%load_ext autoreload
%autoreload 2
%load_ext line_profiler
In [2]:
import scanpy as sc
import random
from unicoord import scu
from unicoord.visualization import draw_loss_curves
import torch
from line_profiler import LineProfiler
In [3]:
sc.settings.verbosity = 3 # verbosity: errors (0), warnings (1), info (2), hints (3)
sc.logging.print_header()
# sc.settings.set_figure_params(dpi=80, facecolor='white')
sc.settings.set_figure_params(vector_friendly=False)
hECA total¶
In [4]:
adata = sc.read_h5ad(r'F:\h5ad\tabularMuris\TBMU.h5ad', )
In [5]:
adata
Out[5]:
In [6]:
adata[adata.obs.cell_ontology_class.isna()].obs.cell_ontology_class[0]
Out[6]:
In [7]:
adata.obs.cell_ontology_class.dtype.name
Out[7]:
In [8]:
for c in adata.obs.columns:
a = adata.obs[c]
if a.dtype.name == 'category':
adata.obs[c] = a.astype(str)
In [31]:
scu.model_unicoord_in_adata(adata,
n_cont=50, n_diff=0, n_clus = [],
obs_fitting=['cell_ontology_class',
'free_annotation','mouse.id','mouse.sex',
'tissue','tissue_tSNE_1','tissue_tSNE_2','seq_tech'],
min_obs = 500)
In [32]:
unc_stuffs = adata.uns['unc_stuffs']
In [33]:
scu.train_unicoord_in_adata(adata, epochs=10, chunk_size=20000, slot = "cur")
In [15]:
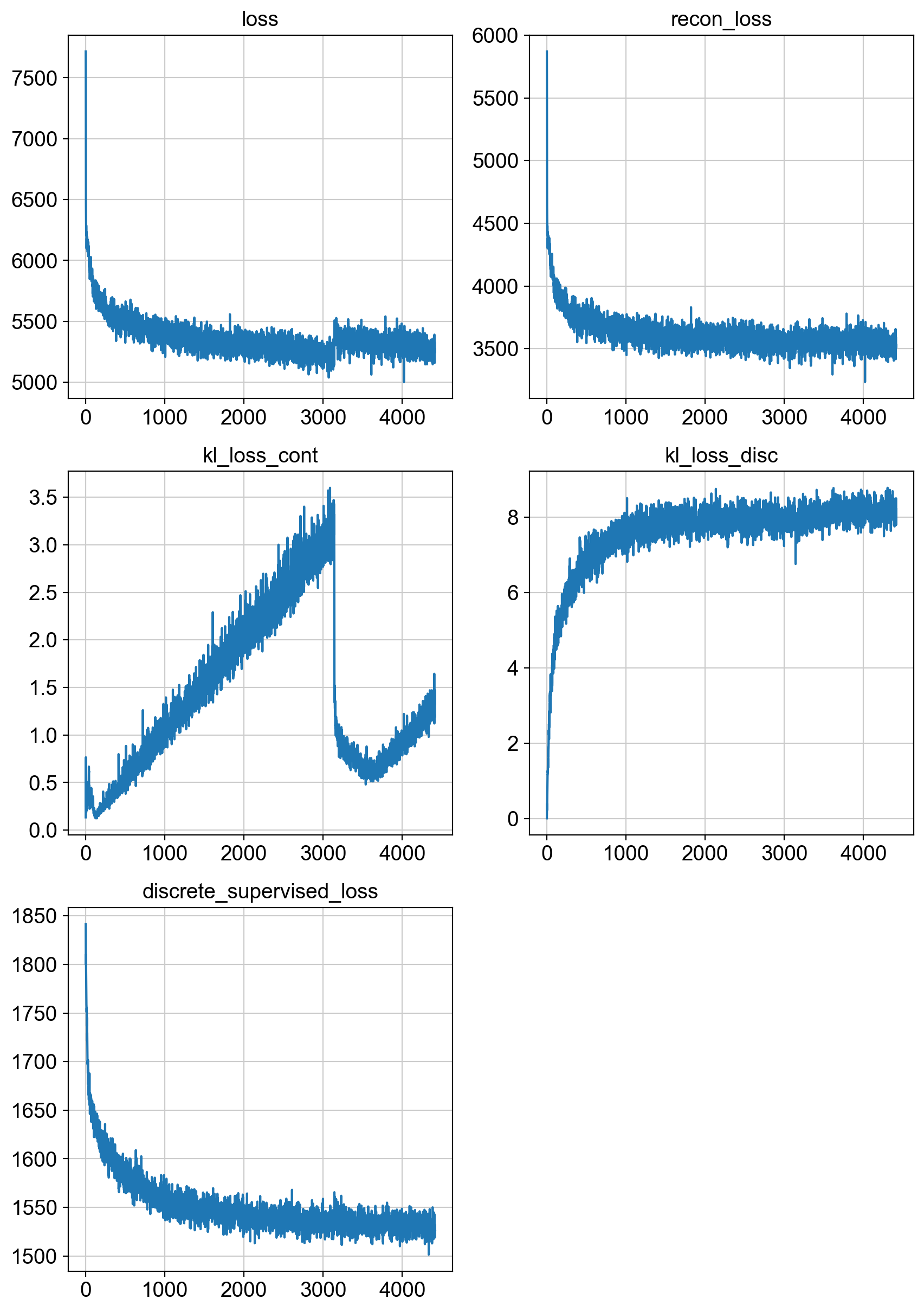
fig = draw_loss_curves(unc_stuffs['loss'])
# if save_figs:
# fig.savefig(os.path.join(savePath, 'img', 'fig1_lossCurves.png'))
fig.show()
In [34]:
scu.write_scu_h5ad(adata, './pretrained_models/TBMU.h5ad', only_model=True)
predict IPF¶
In [9]:
cdata = scu.load_scu_h5ad('./pretrained_models/TBMU.h5ad')
In [61]:
bdata = sc.read_h5ad('./IPF.h5ad')
In [62]:
bdata = bdata.raw.to_adata()
sc.pp.normalize_total(bdata, target_sum=1e4 ,exclude_highly_expressed= True)
sc.pp.log1p(bdata)
bdata
Out[62]:
In [63]:
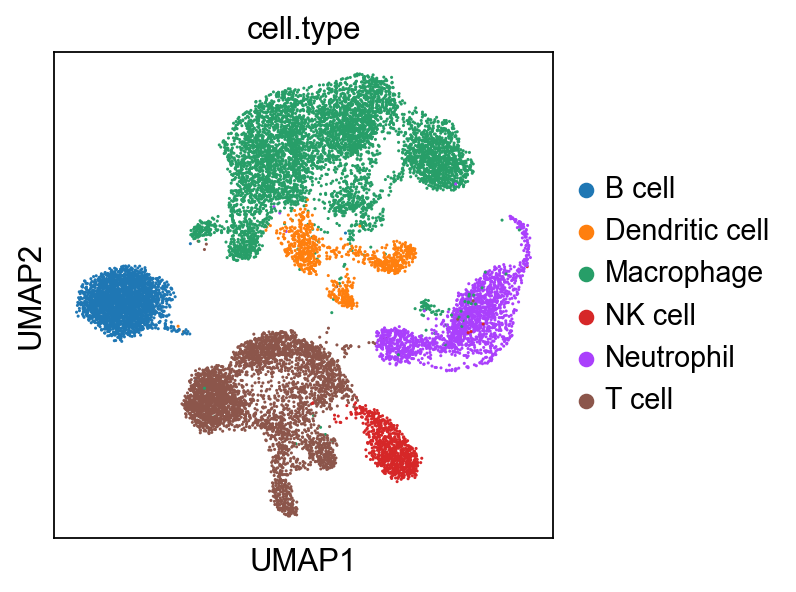
ct_dict = {0:'Macrophage',1:'Dendritic cell',2:'T cell',3:'B cell',4:'Neutrophil',5:'NK cell'}
bdata.obs['cell.type'] = [ct_dict[i] for i in bdata.obs['cell.type']]
In [64]:
sc.pl.umap(bdata, color=['cell.type'])
In [65]:
scu.predcit_unicoord_in_adata(bdata, cdata)
In [66]:
sc.pl.umap(bdata,
color=['cell.type']+[s+'_unc_infered' for s in \
['cell_ontology_class','mouse.id','mouse.sex','tissue','seq_tech']],
ncols = 1)
In [60]:
bdata.obs.cell_ontology_class_unc_infered.value_counts()
Out[60]:
In [71]:
bdata.obs.groupby(['cell_ontology_class_unc_infered','cell.type']).size().unstack().to_csv('./TBMU_ct_pred.csv')
predict it self¶
In [10]:
scu.predcit_unicoord_in_adata(adata, cdata)
In [11]:
torch.cuda.empty_cache()
In [12]:
adata
Out[12]:
In [15]:
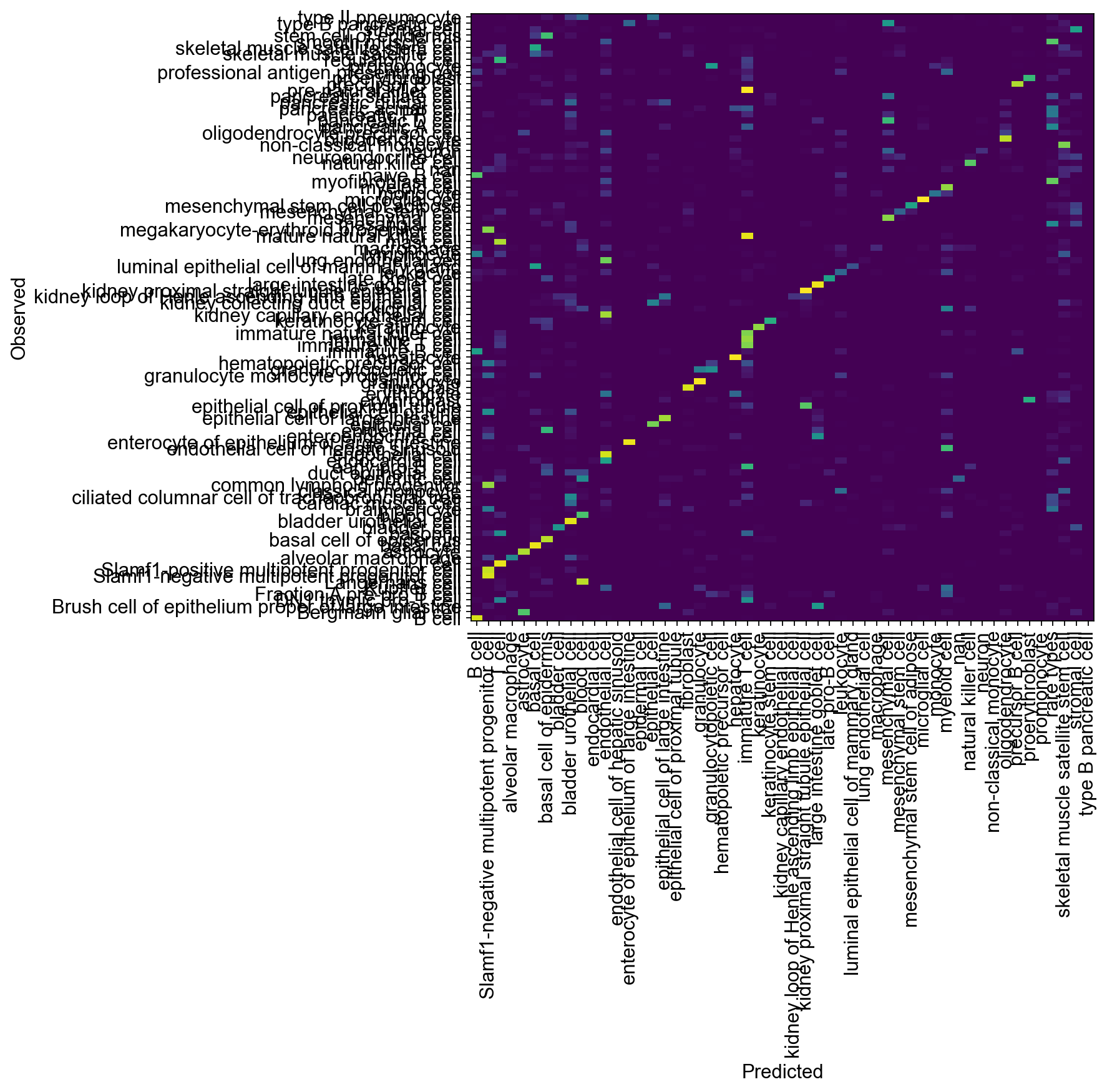
df = adata.obs.groupby(["cell_ontology_class",
"cell_ontology_class_unc_infered"]).size().unstack(fill_value=0)
conf_mat = df / df.sum(axis=1).values[:, np.newaxis]
In [19]:
df.to_csv('./TBMU_ct_self.csv')
In [17]:
import matplotlib.pyplot as plt
In [18]:
plt.figure(figsize=(8, 8))
_ = plt.pcolor(conf_mat)
_ = plt.xticks(np.arange(0.5, len(df.columns), 1), df.columns, rotation=90)
_ = plt.yticks(np.arange(0.5, len(df.index), 1), df.index)
plt.xlabel("Predicted")
plt.ylabel("Observed")
Out[18]:
In [16]:
df
Out[16]: